Division of Molecular Biology & Human Genetics
Tuberculosis Host GeneticsResearch
Human Genetic Diversity
 Genetic
variation between humans can be ascribed to differences between
individuals within populations (85-90%) and to differences between
populations (10-15%). As humans migrated out of Africa, they adapted to
new environments and groups became isolated from one another. This
resulted in different frequencies of genetic variants in the resultant
populations.
Genetic
variation between humans can be ascribed to differences between
individuals within populations (85-90%) and to differences between
populations (10-15%). As humans migrated out of Africa, they adapted to
new environments and groups became isolated from one another. This
resulted in different frequencies of genetic variants in the resultant
populations.Evidence from mitochondrial DNA and Y-chromosome
studies shows that the San of Southern Africa are likely to be the
oldest human population group. We have participated in a collaboration
with researchers from the USA to collect samples from this population
group for genome-wide analysis, which will be an important resource for
gaining insight into the genetic history of humans.
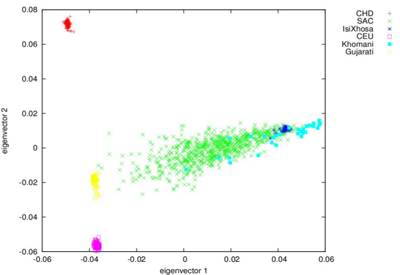
Admixture
occurs when two or more previously separated population groups produce
offspring. The predominant population group in the Western Cape, South
Africa, is the admixed group known officially as the South African
Coloured (SAC). The SAC had their origins in the diverse groups in the
early days of Cape history, including Europeans settlers, the slaves
they brought in from Indonesia, India and other parts of Africa, local
Bantu-speakers, and the indigenous Khoe-San. They therefore constitute a
complex combination of continental populations.
We are using the
powerful technique of admixture mapping to find genetic variants that
differ in frequency between source populations of the SAC and may have a
role in susceptibility to tuberculosis.
In addition, proportions
of ancestry received from source populations vary between individuals in
the population and this may have a confounding effect in genetic
association studies if the differences are not homogeneous between case
and control groups. We have therefore also developed a small panel of
ancestry informative markers (AIMs) that can be used to adjust for
admixture in candidate gene association studies of the SAC – see
Software.

